Landmark_reference_pt_dist¶
This is a function to measure the distance from user defined points to the centroid and a point defined by the centroid coordinate along the x-axis and baseline coordinate (top of pot) along the y-axis. Calculating the vertical distance between leaf tip points to the centroid of the plant object in side-view images may provide a proxy measure of turgor pressure.
plantcv.landmark_reference_pt_dist(points_r, centroid_r, bline_r)
returns none
- Parameters:
- points_r - A list of tuples representing rescaled landmark points
- centroid_r - A tuple representing the rescaled centroid point
- bline_r - A tuple representing the rescaled baseline point
- Context:
- Used to estimate the distance and angles of landmark points relative to shape reference landmarks (centroid and pot height aka baseline)
- Output data stored: Data ('vert_ave_c', 'hori_ave_c', 'euc_ave_c', 'ang_ave_c', 'vert_ave_b', 'hori_ave_b', 'euc_ave_b',
'ang_ave_b') automatically gets stored to the
Outputsclass when this function is ran. These data can always get accessed during a workflow (example below). For more detail about data output see Summary of Output Observations
Input rescaled points, centroid and baseline points
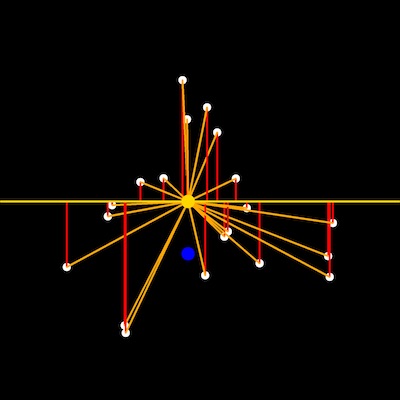
from plantcv import plantcv as pcv
# Set global debug behavior to None (default), "print" (to file),
# or "plot" (Jupyter Notebooks or X11)
pcv.params.debug = "print"
# Identify acute vertices (tip points) of an object
# Results in set of point values that may indicate tip points
pcv.landmark_reference_pt_dist(points_r, centroid_r, bline_r)
# Access data stored out from landmark_reference_pt_dist
avg_vert_distance = pcv.outputs.observations['vert_ave_c']['value']
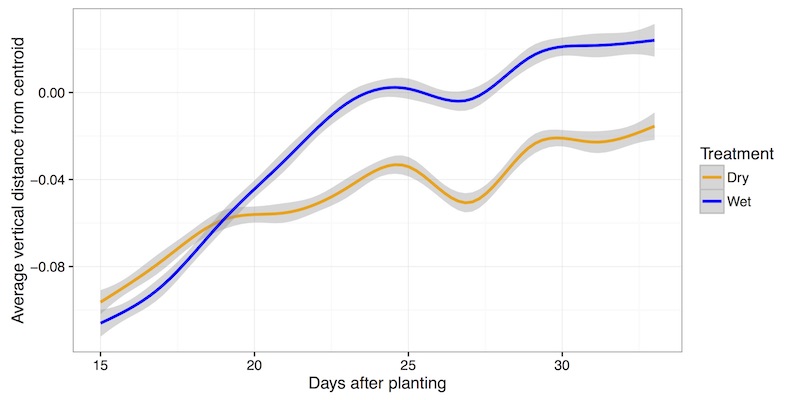
Representation of many data points collected in two treatment blocks throughout time
Source Code: Here