Analyze grayscale values¶
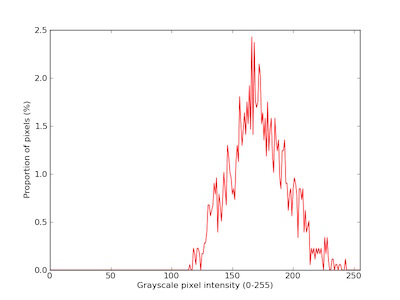
This function calculates the intensity of each pixel associated with the plant and writes the values out to the Outputs class. Can also return/plot/print out a histogram plot of pixel intensity.
plantcv.analyze.grayscale(gray_img, labeled_mask, n_labels=1, bins=100, label=None)
returns Histogram image
- Parameters:
- gray_img - 8- or 16-bit grayscale image data.
- labeled_mask - Labeled mask of objects (32-bit).
- n_labels - Total number expected individual objects (default = 1).
- bins - Number of histogram bins (default = 100)
- label - Optional label parameter, modifies the variable name of observations recorded. Can be a prefix or list (default = pcv.params.sample_label).
- Context:
- Grayscale pixel values within a masked area of an image.
-
Example use:
-
Output data stored: Data ('gray_frequencies', 'gray_mean', 'gray_median', 'gray_stdev') automatically gets stored to the
Outputsclass when this function is ran. These data can always get accessed during a workflow (example below). For more detail about data output see Summary of Output Observations
Original grayscale image

from plantcv import plantcv as pcv
# Set global debug behavior to None (default), "print" (to file),
# or "plot" (Jupyter Notebooks or X11)
pcv.params.debug = "plot"
# Optionally, set a sample label name
pcv.params.sample_label = "plant"
# Caclulates the proportion of pixels that fall into a signal bin and writes the values to a file.
# Also provides a histogram of this data
analysis_image = pcv.analyze.grayscale(gray_img=gray_img, labeled_mask=mask, n_labels=1, bins=100)
# Access data stored out from analyze.grayscale
nir_frequencies = pcv.outputs.observations['plant_1']['gray_frequencies']['value']
Near-infrared signal histogram
Source Code: Here