Fill Segments¶
Propagate the labels of a segmented skeleton to fill the mask.
plantcv.morphology.fill_segments(mask, objects, stem_objects=None, label=None)
returns filled_mask
- Parameters:
- mask - Binary mask
- objects - Segment objects (output from either plantcv.morphology.prune, plantcv.morphology.segment_skeleton, or plantcv.morphology.segment_sort).
- stem_objects - Optional input for stem objects that will be combined into a single object before filling the mask.
- label - Optional label parameter, modifies the variable name of observations recorded. (default =
pcv.params.sample_label)
- Context:
- Uses the watershed algorithm to fill the mask propagating the objects' labels.
- Output data stored: Data ('segment_area') automatically gets stored to the
Outputsclass when this function is ran without thestem_objectsparameter. When the optional parameter is utilized then the data variable names will be ('leaf_area') and ('stem_area'). Data sample names are modified with an optionallabelprefix. These data can always get accessed during a workflow (example below). For more detail about data output see Summary of Output Observations
Reference Image: mask, objects drawn as labels

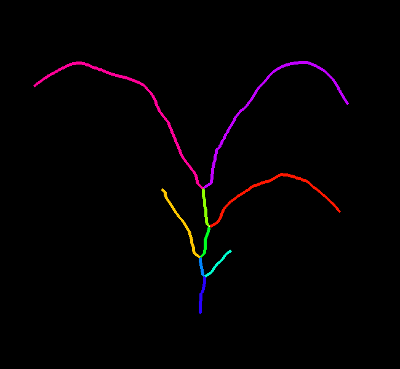
from plantcv import plantcv as pcv
# Set global debug behavior to None (default), "print" (to file),
# or "plot" (Jupyter Notebooks or X11)
pcv.params.debug = "plot"
# Optionally, set a sample label name
pcv.params.sample_label = "plant"
filled_mask = pcv.morphology.fill_segments(mask=plant_mask, objects=obj)
# Convert labeled mask to a colorized image
filled_image = pcv.visualize.colorize_label_img(label_img=filled_mask)
# Access data stored out from fill_segments
segments_area = pcv.outputs.observations['plant']['segment_area']['value']
Filled Image

Source Code: Here